Glossary¶
- Aorta¶
The aorta is the largest artery of the body and carries blood from the heart to the circulatory system. It has several sections: The Aortic Root is the transition point where blood first exits the heart. It functions as the water main of the body. The Aortic arch, is the curved segment that gives the aorta its cane-like shape. It bridges the ascending and descending aorta.
- Ascending Aorta¶
The ascending aorta is the first part of the aorta, which is the largest blood vessel in the body. It comes out of your heart and pumps blood through the aortic arch and into the descending aorta.
- Descending Aorta¶
The descending aorta is the longest part of your aorta (the largest artery in your body). It begins after your left subclavian artery branches from your aortic arch, and it extends down into your belly. The descending aorta runs from your chest (thoracic aorta) to your abdominal area (abdominal aorta).
- Organ Segmentation¶
The definition of the organ boundary or organ segmentation is helpful for the orientation and identification of the regions of interest inside the organ during the diagnostic or treatment procedure. Further, it allows the volume estimation of the organ, such as the aorta.
- inferior¶
Inferior is the direction away from the head; the lower (e.g., the foot is part of the inferior extremity).
- superior¶
Superior is the direction toward the head end of the body; the upper (e.g., the hand is part of the superior extremity).
- slice¶
A 2-dimensional image is retrieved from a 3-dimensional volume.
- kernel size¶
The size of the kernel for binary dilation. The binary dilation section shows an example of binary dilation with a kernel size of 3.
- label map¶
A labeled map or a label image is an image that labels each pixel of a source image. For example, the time zone map shown below has 4 labels to label the time zone for each state in the U.S.
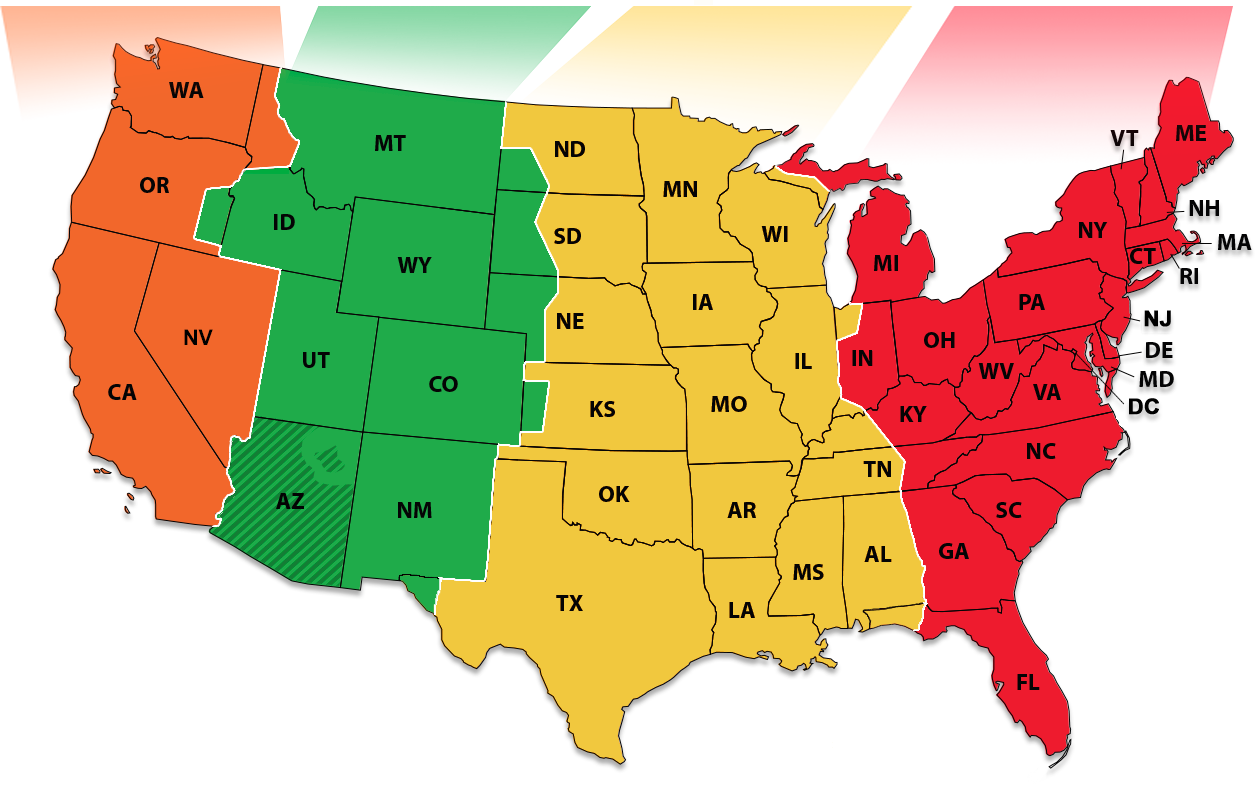
Fig 1. U.S. time zone map with 4 labels.¶
- binary dilation¶
Binary dilation is a mathematical morphology operation that uses a structuring element (kernel) for expanding the shapes in an image. Using scipy.binary_dilation as example:
>>> from scipy import ndimage >>> a array([[ 0., 0., 0., 0., 0.], [ 0., 0., 0., 0., 0.], [ 0., 0., 1., 0., 0.], [ 0., 0., 0., 0., 0.], [ 0., 0., 0., 0., 0.]]) >>> # the default kernel shape of scipy is a 3x3 structuring element with connectivity 1 >>> ndimage.binary_dilation(a).astype(a.dtype) array([[ 0., 0., 0., 0., 0.], [ 0., 0., 1., 0., 0.], [ 0., 1., 1., 1., 0.], [ 0., 0., 1., 0., 0.], [ 0., 0., 0., 0., 0.]])
- rms_error¶
Value of RMS change below which the filter should stop. This is a convergence criterion.
- Maximum iteration¶
Number of iterations to run
- Curvature scaling¶
Weight of the curvature contribution to the speed term.
- Propagation scaling¶
Weight of the propagation contribution to the speed term.
- segmented slice¶
A 2-dimensional image retrieved by applying SITK::ThresholdSegmentationLevelSetImageFilter with the euclidean distance transform image, the original image, and the threshold value calculated with the mean and the standard deviation of the intensity values that were labeled as the white pixel.
- contour line¶
A contour line (also isoline, isopleth, or isarithm) of a function of two variables is a curve along which the function has a constant value so that the curve joins points of equal value.
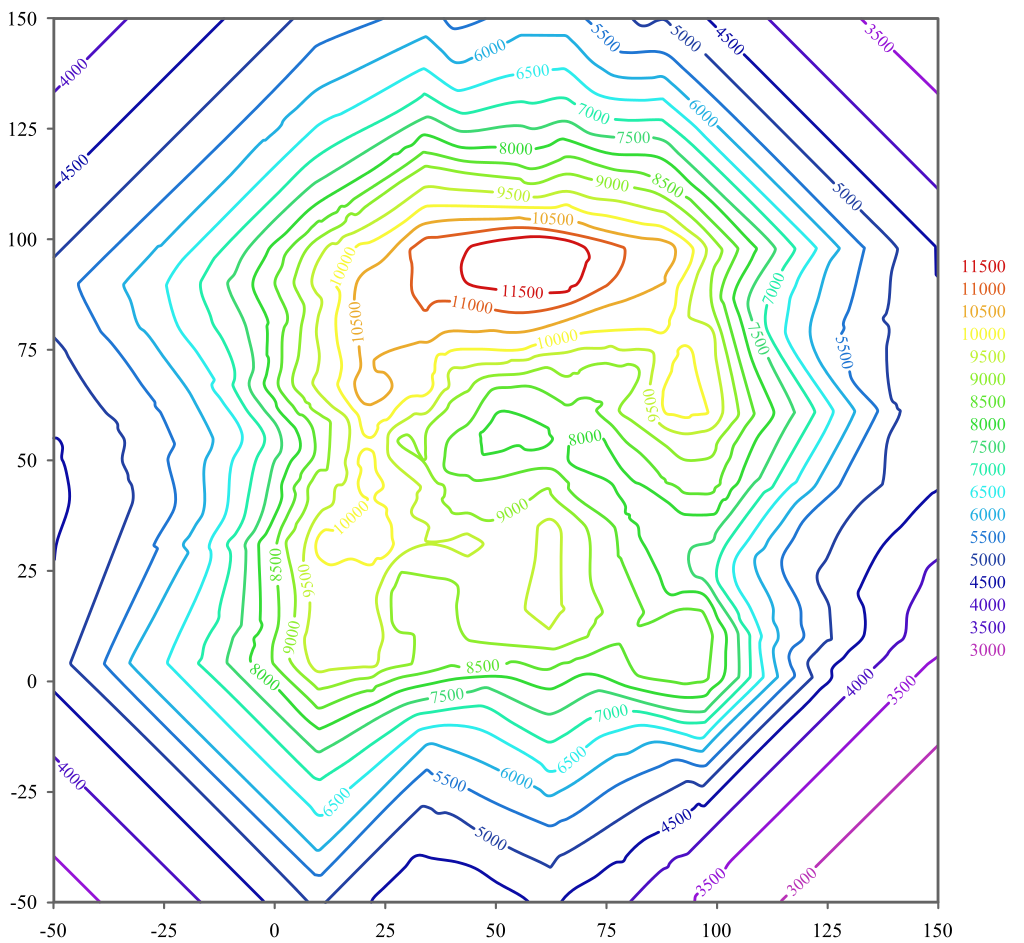
Fig 2. A contour line map¶
- Level sets¶
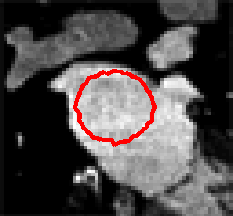
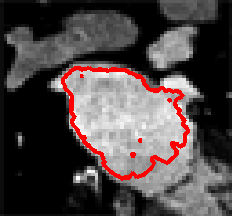
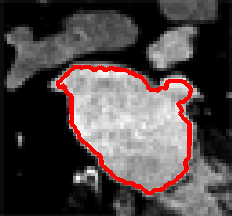
Level Sets are an important category of modern image segmentation techniques based on partial differential equations (PDE), i.e. progressive evaluation of the differences among neighboring pixels to find object boundaries. The pictures below demonstrate an example of how Level Sets method work on finding the region of the heart. It starts with a seed contour that is within the region of interest, then by finding the gradient based on the contour line, the segmentation result will propagate towards outside of the region until the maximum difference between the neighboring pixels are reached.
- threshold¶
A threshold is an amount, level, or limit on a scale. When the threshold is reached, something else happens or changes. For SITK::ThresholdSegmentationLevelSetImageFilter, this inputs decides whether a new pixel should be included in the segmentation region or not based on the threshold intensity values. The algorithm needs a lower and upper threshold.
- threshold coefficient¶
This coefficient is used to compute the lower and upper threshold passing through the segmentation filter SITK::ThresholdSegmentationLevelSetImageFilter. The algorithm first uses SITK::LabelStatisticsImageFilter to get the mean and the standard deviation of the intensity values of the pixels that are labeled as the white pixel. Larger values with this coefficient imply a larger range of thresholds when performing the segmentation, which leads to a larger segmented region.
Note
The lower threshold is calculated as the mean of the intensity values subtract from the threshold coefficient multiplied by the standard deviation. The upper threshold is calculated by adding the threshold multiplied by the standard deviation.
- Stop limit¶
This limit is used to stop the segmentation algorithm. It is used differently in segmentation in inferior direction and segmentation in superior direction.
In inferior direction, if the new centroid that is closest to the previous ascending aorta centre is still reaching the distance limit, then the algorithm will stop consider taking the new centroid closer to the ascending aorta. In other words, only 1 centroid will be used for descending aorta segmentation.
In supeiror direction, if the standard deviation of the inital label map and the segmentation result label map has larger difference than the limit, the algorithm will stop processing segmentation for the rest of the slices. For example, assume that the standard deviation of the initial label image is 20, and the standard deviation of the segmentation label image is 40, with stop limit of 10, the program hault immediately.
- Euclidean distance transform¶
The euclidean distance transform is the map labeling each pixel of the image with the distance to the nearest obstacle pixel (black pixel for this project). SITK::SignedMaurerDistanceMapImageFilter will also reverse propagate the obstacle pixel by setting them to a lower values (the outmost obstacle pixel will have the lowest value.)
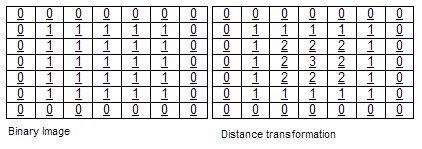
The euclidean distance transform image¶
- DICOM¶
Digital Imaging and Communications in Medicine (DICOM) is the standard for the communication and management of medical imaging information and related data.
- 3D Slicer¶
3D Slicer (Slicer) is a free and open source software package for image analysis and scientific visualization. Slicer is used in a variety of medical applications, including autism, multiple sclerosis, systemic lupus erythematosus, prostate cancer, lung cancer, breast cancer, schizophrenia, orthopedic biomechanics, COPD, cardiovascular disease, and neurosurgery.